McMaster researchers use AI tech to unveil key mechanism of bacterial warfare
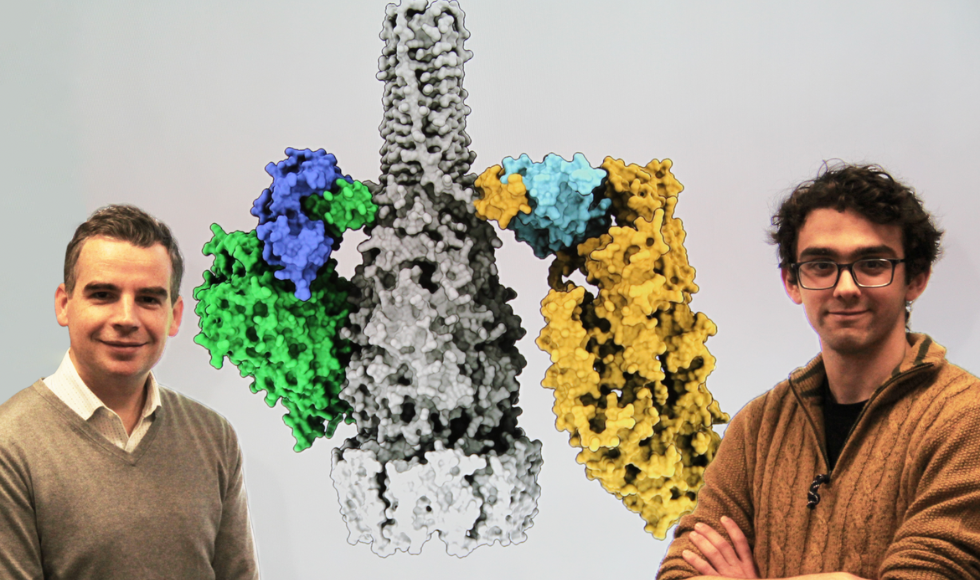
John Whitney (left) and Jake Colautti (right) showcase a 3D rendering of a bacterial secretion system (black and white) and its associated “adaptor proteins” (blues). These proteins, the research team says, allow bacteria to secrete potent toxins (yellow/green) — a key mechanism of bacterial warfare.
BY Blake Dillon
December 19, 2024
Bacteria are in a constant fight for survival, and they rely on an array of deadly weapons to keep their competitors at bay. One such weapon is a sophisticated secretion system, which allows certain bacteria — some that cause disease and some that don’t — to inject toxic proteins into rival cells.
While this system is becoming increasingly well-studied, how it selects diverse proteins for secretion has effectively remained a mystery — until now.
In a new study published in the journal Nature Communications, McMaster University researchers leveraged AI to detail the structure and function of the proteins involved in this complex secretion process.
“This system is present in a broad range of bacteria and plays a key role in inter-microbial warfare,” says John Whitney, an associate professor in McMaster’s Department of Biochemistry & Biomedical Sciences and lead investigator on the study. “It’s been known for some time that it can recognize and secrete different proteins, but we wanted to know exactly how it’s able to do so.”
Just like computer adaptors
Jake Colautti, a graduate student in Whitney’s lab and the lead author on the new paper, says that this system secretes a small but specific set of toxins that are pulled from a pool of thousands of different proteins. But extreme precision is not what sparked this study — rather, that those toxic proteins that are selected are seemingly unrelated, both structurally and evolutionarily.
This is a stark contrast to other bacterial secretion systems, which, researchers say, have shown to eject toxins that share common traits. For instance, earlier this year, Whitney’s team described a barcode-like characteristic present in another similar system, which draws a clear connection between the toxins that are secreted.
But there are no barcodes here, says Whitney, a member of the Michael G. DeGroote Institute for Infectious Disease Research.
Instead, researchers found something a lot more subtle: a family of intermediary “adaptor proteins” that bind to the secretion system and allow it to select and secrete a range of distinct and unrelated toxins.
As the name implies, the researchers say that these proteins are similar to computer adaptors — a single attachment that enables compatibility with a broad range of accessories. In this case, the accessories aren’t HDMI cables or keyboards — they’re toxic proteins that are pumped out to kill competitors, including host cells and other bacteria.
“These proteins are exactly like computer adaptors, in that they have one end that is shared across the family, and one end that is variable,” says Colautti. “That variable end has evolved over millions of years to recognize specific toxins.”
Turning to AI for answers
To study these adaptor proteins, researchers had to first detail their physical shape — “in the protein world, structure is function,” says Colautti.
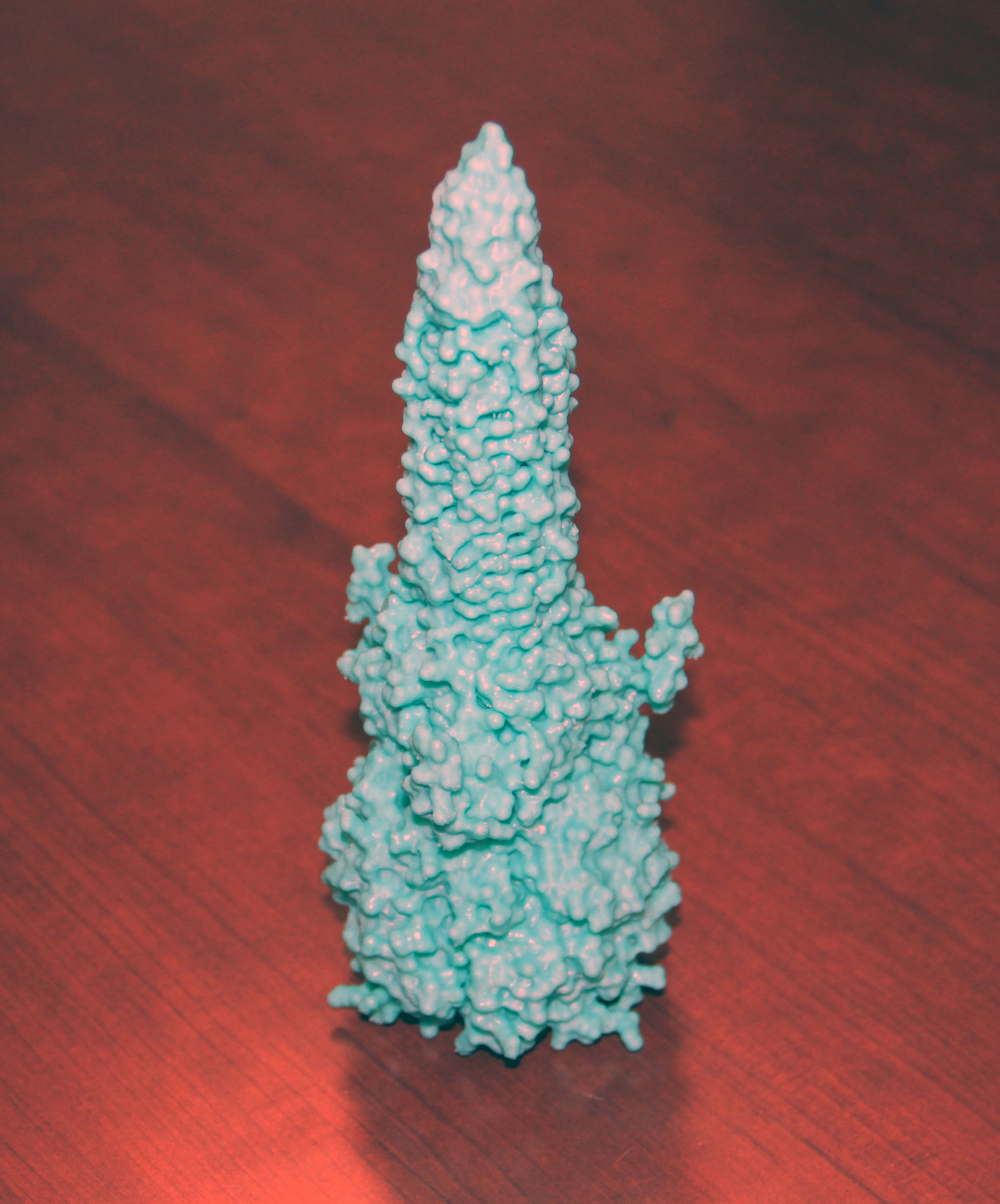
Gesturing to a large 3D-printed model of the secretion system’s spike protein (pictured), he suggests that most people, even without prior knowledge, might guess that its function is to stab into other cells, given its pointy shape — “and they’d be right,” he says.
Colautti says this concept applies broadly — that understanding what proteins look like can often be the first clue for what they might do. The problem, though, is that detailing the physical structure of these microscopic proteins can take years, making it challenging for scientists to efficiently infer their functions.
So, for this study, Whitney’s group leaned on an AI program called AlphaFold to fast-track that process. They fed the algorithm prompts to help it predict the complex structure of the system’s spike protein, and then verified those predictions in the lab.
“The AI was right,” Whitney says.
As it has on other fields, Whitney says the introduction of AI into structural biology is having a seismic impact. In fact, the developers of AlphaFold received a 2024 Nobel Prize for the significance of their algorithm, which has already characterized a range of proteins critical to the development of new vaccines, pharmaceuticals, and other health interventions.
“Wet labs aren’t going anywhere, but AI-driven hypotheses are literally saving researchers years,” Whitney says.
Moonlighting in the lab
For Colautti, the time saved was critical to advancing this discovery, given the demands of his busy schedule. Part of McMaster’s MD/PhD Program, a prestigious small-cohort academic pathway for aspiring physician-scientists, he conducted much of this research at night after long days in medical school.
But Colautti says bouncing back and forth between the lab and the clinic keeps things fresh and has given him a greater appreciation for how basic research, like this recent study, can impact patients and vice versa.
“There’s lots of opportunity to bring basic science into the clinic, but being a physician also presents opportunity to encounter interesting biological problems out in the real world,” he says. “I want to take those interesting problems from the clinic to the lab and study them to help answer questions that improve our ability to treat disease.”
Meredith Vanstone, an associate professor in McMaster’s Department of Family Medicine and the director of the MD/PhD Program, says the program is designed to foster exactly that kind of mindset.
“Physician scientists have a unique vantage point that allows them to identify important problems and conceptualize and lead the research that will help solve them,” she says, noting that students in the program are encouraged to keep one foot in both worlds throughout the duration of their studies, so that they can truly harness that unique perspective.
Colautti says doing just that helped him see the clinical implications of his recent study, which he believes could eventually inform follow-on research into drug discovery or antimicrobial resistance.